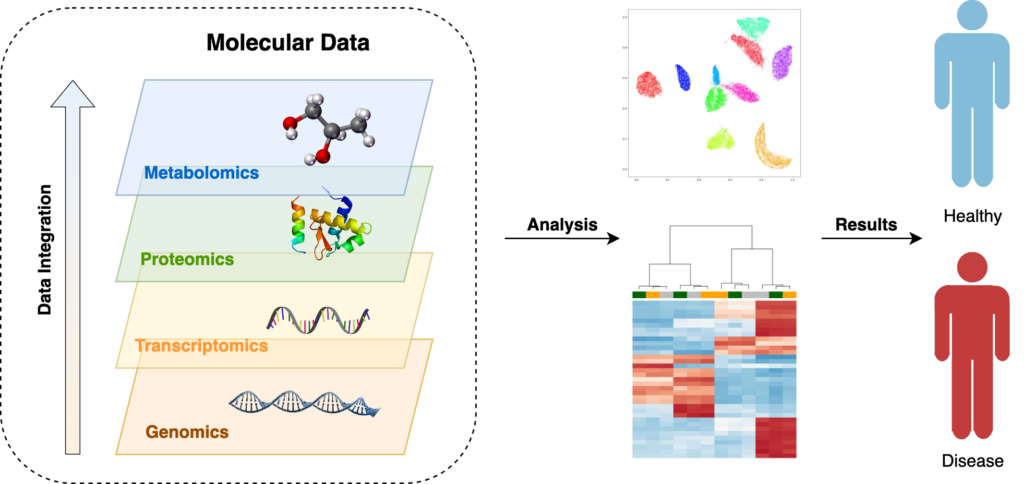
Multi-omics is a rapidly growing field that involves the simultaneous analysis of multiple omics data types, such as genomics, transcriptomics, proteomics, and metabolomics. The integration of multi-omics data enables a more comprehensive understanding of biological systems, leading to new discoveries and insights into the underlying mechanisms of diseases. The importance of bioinformatics in multi-omics research cannot be overstated, as it plays a critical role in the analysis, interpretation, and visualization of these complex data sets.
The use of multi-omics has several advantages over traditional single-omics approaches. By analyzing multiple omics data types, researchers can identify biological pathways and networks that are disrupted in disease, leading to the identification of new drug targets and therapeutic approaches. For example, multi-omics analysis has been used to identify biomarkers for cancer and develop personalized therapies for patients.
The analysis of multi-omics data requires the integration of different bioinformatics tools and techniques, such as statistical analysis, machine learning, and network analysis. These tools enable researchers to identify patterns and relationships between different omics data types and extract meaningful information from the vast amount of data generated. The integration of multi-omics data also requires standardized data formats and ontologies, which facilitate data sharing and collaboration between different research groups.
While multi-omics analysis holds great potential, it can pose significant challenges, especially for those new to the field. Here, we outline a few obstacles that we have encountered during our own analyses:
- Data integration: Integrating diverse omics data sets with varying formats, resolutions, and sources is a significant challenge. Each omics data type has its own complexities and biases, and harmonizing these data sets to extract meaningful information requires robust computational methods and bioinformatics tools.
- Statistical and computational methods: Developing appropriate statistical and computational models to analyze multi-omics data is a significant challenge. Traditional statistical approaches may not be suitable for integrating multiple data types, and novel methods are required to account for complex interactions and dependencies between different omics layers.
- Biological interpretation and validation: Extracting meaningful biological insights from multi-omics data is a complex task. Integrating different layers of omics data can provide a holistic view of biological processes, but it also requires expertise in diverse biological domains and the ability to validate findings using experimental approaches.
The analysis of multi-omics data has numerous applications in different fields, including personalized medicine, drug discovery, and systems biology. By integrating different omics data types, researchers can identify biomarkers for disease, predict treatment response, and develop new therapeutic strategies. Multi-omics analysis can also reveal the complex interplay between different biological systems, leading to a better understanding of the underlying mechanisms of diseases.
Biology is a multidimensional puzzle, and data integration is the tool that helps us piece together the intricate relationships between genes, phenotypes, and environmental factors.
In summary, multi-omics is a powerful approach that enables a more comprehensive understanding of biological systems and diseases. The integration of different omics data types requires the use of bioinformatics tools and techniques, which play a critical role in the analysis, interpretation, and visualization of these complex data sets. The applications of multi-omics analysis are diverse, ranging from drug discovery to systems biology, and have the potential to revolutionize our understanding of biological systems and lead to new therapeutic approaches.